Diel Cycle
Microbial communities in the tropical air ecosystem follow a precise diel cycle
 (SASS® 3100 high-volume air samplers located at Nanyang Technological University (NTU) campus)
(SASS® 3100 high-volume air samplers located at Nanyang Technological University (NTU) campus)
The last unexplored ecosystem
 (Birds eye view of sampling location at Nanyang Technological University (NTU) campus)
The atmosphere is vastly underexplored as a habitable ecosystem for microbial organisms. Of the 3 planetary ecosystems, the lithosphere, hydrosphere, and atmosphere, only soil and aquatic environments have been extensively investigated for their microbial content. In contrast, due to its breadth and paucity of microorganisms, the study of the atmosphere as a microorganism-harbouring ecosystem has been technologically challenging, rendering it an understudied biosphere. The atmosphere has long been assumed to contain microorganisms from local and remote sources, being dispersed for as long as they remain suspended in air. Previously reported numbers of cells found in a defined volume of air have been based on cultivation (colony-forming units; CFUs) or estimated by amplified ribosomal molecular markers and is reported to be ∼102 cells/L of air sampled (compared to the microbial cellular density of ∼1012 cells/L of soil and ∼109 cells/L of water). This 7 to 10 log difference in cell density per volume represents a significant technological challenge for analyzing the biological content of air, as it mandates very large volumes to be collected over what is often extended periods of time from days to months.
(Birds eye view of sampling location at Nanyang Technological University (NTU) campus)
The atmosphere is vastly underexplored as a habitable ecosystem for microbial organisms. Of the 3 planetary ecosystems, the lithosphere, hydrosphere, and atmosphere, only soil and aquatic environments have been extensively investigated for their microbial content. In contrast, due to its breadth and paucity of microorganisms, the study of the atmosphere as a microorganism-harbouring ecosystem has been technologically challenging, rendering it an understudied biosphere. The atmosphere has long been assumed to contain microorganisms from local and remote sources, being dispersed for as long as they remain suspended in air. Previously reported numbers of cells found in a defined volume of air have been based on cultivation (colony-forming units; CFUs) or estimated by amplified ribosomal molecular markers and is reported to be ∼102 cells/L of air sampled (compared to the microbial cellular density of ∼1012 cells/L of soil and ∼109 cells/L of water). This 7 to 10 log difference in cell density per volume represents a significant technological challenge for analyzing the biological content of air, as it mandates very large volumes to be collected over what is often extended periods of time from days to months.
 (Sets of 3-samplers set-up for cascade time delay at 2-hour intervals)
Research into airborne microbial organisms has been predominantly carried out in countries located within temperate climate zones or as global surveys with varying geographic and climatic parameters. As a tropical, equatorial country, Singapore is located in the intertropical convergence zone and experiences a biannual reversal of wind directions, which defines the 2 monsoon seasons. The biannually alternating wind directions of the study site could result in vastly disparate sources of transported microorganisms helping to define and answer the question of local or remote sources. The small variation in length of daylight across the seasons as well as absence of large day-to-day fluctuations in temperature and relative humidity are factors making the tropical region an ideal air microbiome research site.
(Sets of 3-samplers set-up for cascade time delay at 2-hour intervals)
Research into airborne microbial organisms has been predominantly carried out in countries located within temperate climate zones or as global surveys with varying geographic and climatic parameters. As a tropical, equatorial country, Singapore is located in the intertropical convergence zone and experiences a biannual reversal of wind directions, which defines the 2 monsoon seasons. The biannually alternating wind directions of the study site could result in vastly disparate sources of transported microorganisms helping to define and answer the question of local or remote sources. The small variation in length of daylight across the seasons as well as absence of large day-to-day fluctuations in temperature and relative humidity are factors making the tropical region an ideal air microbiome research site.
Here, we investigated the microbial composition of the outdoor air microbiome at a single site in the tropics (Singapore, N1.334731; E103.680996, sea-surface level) through a metagenomics time course study of unprecedented temporal and taxonomic resolution. Using deep metagenomic sequencing and analysis, we were able to describe the air microbiome variation within time intervals of hours, days, and up to months.
Resolution of the tropical air microbiome – a microbial clock
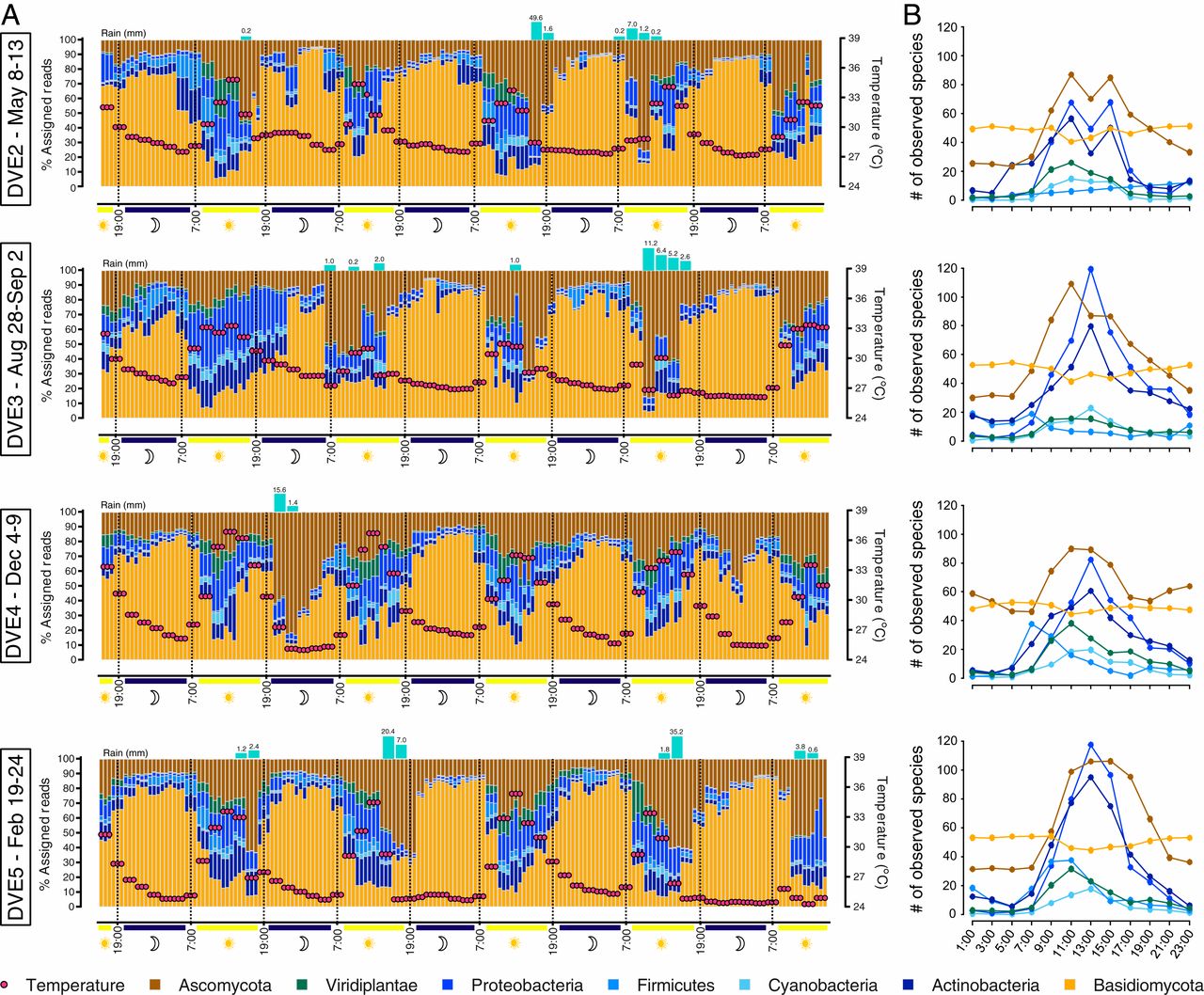
(Taxonomical breakdown and temporal resolution of the air microbial communities. (A) Relative abundance of 7 taxonomic groups of organisms in air. Rain events with recorded amounts of precipitation (blue bars) and temperature distribution (red buttons) are denoted for each time series. The timescale is indicated (Bottom), as well as day and night samples denoted by sun and moon symbols. (B) Observed number of species by taxonomical groups in each of the 4 DVEs. The graph depicts the change in species abundance per group, indicating the robustness of the Basidiomycota community versus strong daily variation of the community composition for the other 6 phyla)
 (On-site sensor collection measuring wind speed, co2, UV, and atmospheric pressure)
In this study, we investigated 795 time-resolved metagenomes from tropical air, generating 2.27 terabases of data, the equivalent of ∼760 human genomes. We have devised an air-sampling approach that allows for 2-h intervals to be sampled over the course of an entire day for one week and we repeated this in 3 month intervals over the course of 13 months.
What we saw was that the airborne microbial organisms followed a clear day-night cycle, where different microbial groups would increase up to ten-fold during midday or during rain events while others became more common during the night. Several environmental factors were considered during sampling such as light (UV), environmental pollutants (SOx/NOx), and wind speed and direction. Though, it was the signature of several specific fungal and bacterial species which were strongly affected by changes in particular factors such as temperature, humidity (including rain events), and CO2 concentration, which would appear to drive this day-night difference. This within-day cyclic pattern was stronger than variation over days, weeks or months. Our environmental time series also revealed the existence of a large core of microbial taxa that remained invariable over 13 months, thereby underlining the long-term robustness of the airborne community structure.
(On-site sensor collection measuring wind speed, co2, UV, and atmospheric pressure)
In this study, we investigated 795 time-resolved metagenomes from tropical air, generating 2.27 terabases of data, the equivalent of ∼760 human genomes. We have devised an air-sampling approach that allows for 2-h intervals to be sampled over the course of an entire day for one week and we repeated this in 3 month intervals over the course of 13 months.
What we saw was that the airborne microbial organisms followed a clear day-night cycle, where different microbial groups would increase up to ten-fold during midday or during rain events while others became more common during the night. Several environmental factors were considered during sampling such as light (UV), environmental pollutants (SOx/NOx), and wind speed and direction. Though, it was the signature of several specific fungal and bacterial species which were strongly affected by changes in particular factors such as temperature, humidity (including rain events), and CO2 concentration, which would appear to drive this day-night difference. This within-day cyclic pattern was stronger than variation over days, weeks or months. Our environmental time series also revealed the existence of a large core of microbial taxa that remained invariable over 13 months, thereby underlining the long-term robustness of the airborne community structure.
Unlike terrestrial or aquatic environments, where prokaryotes are prevalent, the tropical airborne biomass was dominated by DNA from eukaryotic phyla. Strikingly, despite only 9 to 17% of the generated sequence data currently being assignable to taxa, the air harboured a microbial diversity that rivals the complexity of other planetary ecosystems.
Setting the foundation
The robustness and precision of our sampling technique , as illustrated by its replicability, has allowed for the undertaking of this experimental time-series survey across days, weeks, and months. In this way we were able to clearly resolve the close interplay of changing environmental aspects within a day and the microbes we breathe, which we may have otherwise missed. The experimental approach and analysis from this study has laid the groundwork to inform our future environmental surveys that will investigate the spatial dynamics and natural variability of airborne microbial communities. By studying the interrelationship of microbiomes from the 3 planetary ecosystems (atmosphere, terrestrial, and aquatic), it can now be shown that temperature is a global driver of microbial community dynamics.Read the manuscript / Forbes news article (Excerpts taken from manuscript abstract and credit to Linh Anh Cat from Forbes magazine)